Data & Statistical Core
Mouse studies sponsored by The Jackson Laboratory Nathan Shock Center (JAX-NSC) produce an enormous amount of data, including phenotypic, genetic, epigenetic, expression profiling and microbiome profiling data. The JAX-NSC Data and Statistical Core addresses a need among preclinical aging researchers studying the genetic factors that underlie lifespan and healthspan for computational, bioinformatic, and statistical expertise. Across 55 pilot projects funded by the JAX-NSC, the Core has supported the national and international aging research community in 14 states and 4 countries.
The Core maintains a research focus on the cutting-edge management and analysis of complex data types via statistical genetics, machine learning, database development, scientific software engineering, data visualization and cloud computing. Our CLIMB LIMS (Rockstep Inc., Portland ME) provides integrated data management and workflow scheduling processes. Our JAX-NSC Data Portal provides a common framework for data access and dissemination for the Center’s lifespan studies. Our QTLViewers provide an interface for genetic analysis which allows easy and in-depth access to genetic mapping details.
The Core specializes in the design and statistical analysis of aging studies with genetic diversity. In contrast to inbred strains, diversity mouse strains and populations feature stable, well-characterized, segregated genetic variation that more accurately reflects the genetic structure of human populations. They offer powerful approaches to study genetically complex traits by enabling high resolution genetic mapping. To learn more about the Diversity Outbred and Collaborative Cross lines, click here.
Core staff include Drs. Gary Churchill and Alison Luciano. Dr. Churchill is the Core Lead. Dr. Luciano is an analyst.

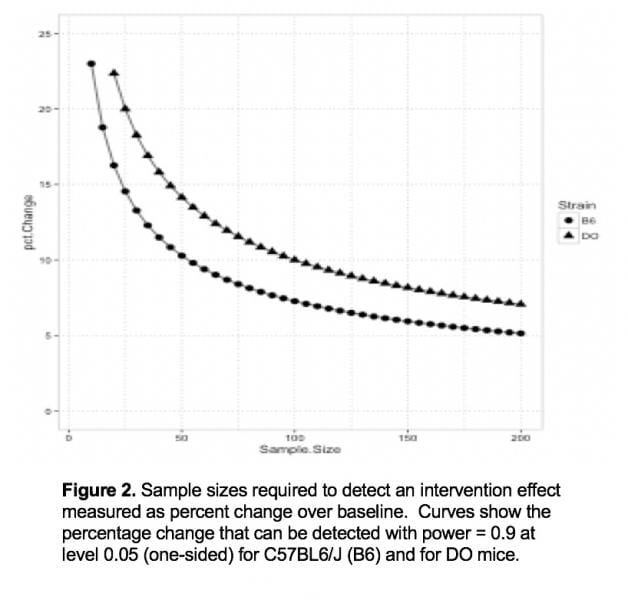
We have established sample size guidelines for intervention studies using DO mice. We computed sample sizes required to detect a change in lifespan between groups of treated and control animals using data from our DO longitudinal study (Figure 2). Due to the added genetic variability in lifespan, DO studies require approximately twice as many animals compared to a C57BL/6J study to detect the same magnitude of change. However, if we consider the possibility that the intervention effect may vary across strains, it makes economic sense to carry out initial studies on the outbred population rather than testing multiple inbred strains.
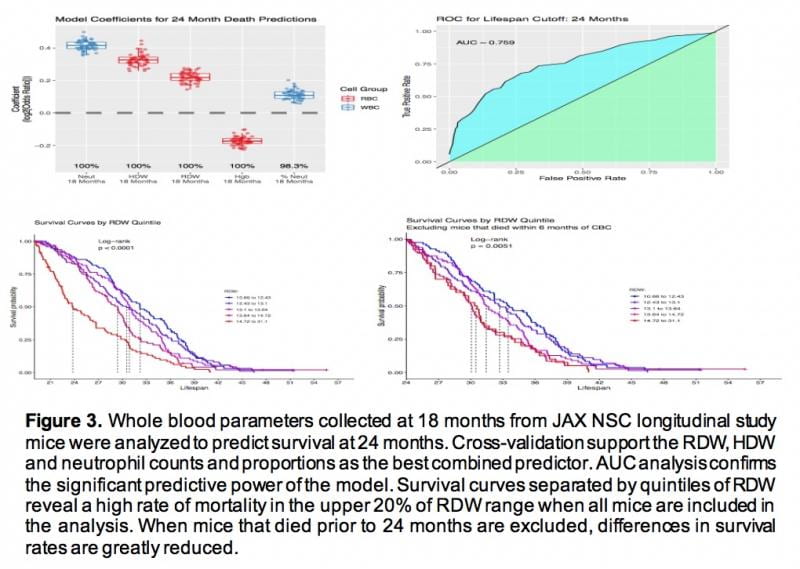
We are evaluating phenotypic (healthspan) data from our longitudinal study to identify potential predictors of lifespan. To date, our findings suggest that long-term predictions (beyond six months) are not better than chance expectation, despite many parameters that show trends with age. However, we can accurately predict death within the next six months (Fig. 3). Findings from the ongoing project suggest that while healthspan para-meters provide reliable indicators of “death’s door,” we have yet to discover the key to long-range prediction of lifespan.